OpenEBench: the ELIXIR platform for benchmarking
Find more information here: https://openebench.readthedocs.io/en/latest/
OpenEBench ( https://openebench.bsc.es ) is the ELIXIR benchmarking and technical monitoring platform for bioinformatics tools, web servers and workflows. OpenEBench is part of the ELIXIR Tools platform and its development is led by the Barcelona Supercomputing Center (BSC) in collaboration with partners within ELIXIR and beyond.
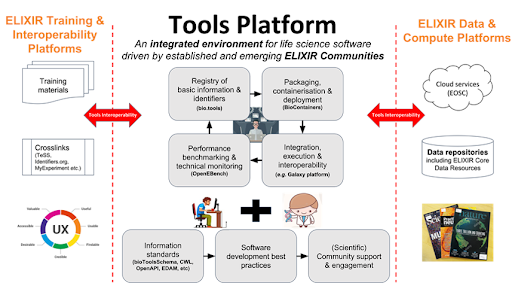
Within the ELIXIR project, OpenEBench is being developed under the Tools Platform at the Work Package 2 (WP2: Benchmarking) ( https://elixir-europe.org/platforms/tools ).
All OpenEBench components have been designed and implemented following the recommendations made by the ELIXIR tools platform e.g. making code available in public repositories from day 1; are available as software containers, and use workflow managers promoted by ELIXIR. Next figure illustrates the interconnection of OpenEBench to other ELIXIR tools platforms systems and platforms and beyond.
The goals of OpenEBench
- Provide guidance and software infrastructure for Benchmarking and Techincal monitoring of bioinformatics tools.
- Engage with existing benchmark initiatives making different communities aware of the platform.
- Maintain a data warehouse infrastructure to keep record of Benchmarking initiatives.
- Expose benchmarking and technical monitoring results to Elixir Tools registry.
- Establish and refine communication protocols with communities and/or infrastructure projects willing to have a unified benchmark infrastructure Coordinate with Elixir.
- Work with the ELIXIR Interoperability Platform to keep FAIR data principles on the Benchmarking data warehouse.
Technical monitoring
OpenEBench, holds a specific infrastructure to monitor software quality. In an initial analysis phase BSC has put together a series of quality metrics taken from a number of sources. The source of such metrics includes documents by the Software Sustainability Institute, recommendations for open source software development, or for software quality metrics. For each metric, a specific source of information have been chosen and the necessary interface implemented.
Scientific benchmarking
Scientific benchmarking helps determine the precision, recall and other metrics of bioinformatics resources in unbiased scenarios, which have been set up through reference databases, ad-hoc input and test data sets reflecting specifying scientific challenges. Chosen metrics allow us to objectively evaluate the relative scientific performance of the different participating resources. It is even possible to understand what are the software potential biases, strengths and weaknesses and/or under which conditions do they perform better or worse.
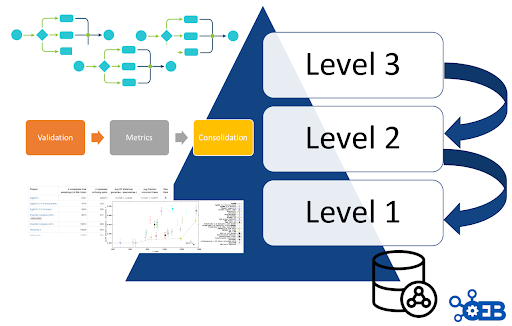
OEB architecture levels
OpenEBench scientific benchmarking architecture has three different levels that allow communities at different maturity stages to make use of the platform.
Level 1
Used for the long-term storage of benchmarking events and challenges aiming at reproducibility and provenance.Level 2
Allows the community to use benchmarking workflows to assess participants’ performance. Those workflows compute one or more evaluation metrics given one or more reference datasets.Level 3
Goes further by getting workflows specifications from participants, and then evaluating them in terms of technical and scientific performance. At this level, the whole benchmarking experiment is performed at OpenEBench; first, the predictions are made using the software provided by the participants, then, those predictions are evaluated with the benchmarking workflows, and, finally, the results are stored and visualized in the web server.
OpenEBench public API's
Although the OpenEBench website gives access to all information stored in the data warehouse in a friendly manner, the platform is designed to provide information in a way that can be integrated in other infrastructures. To this end a series of RESTful API’s have been developed. Information from these APIs is obtained in JSON format.
- OpeneBench tools API: https://openebench.bsc.es/monitor/tool
- OpeneBench scientific API: https://openebench.bsc.es/sciapi/
The white paper for this project is available here